Research Focus
The goal of my research program is to understand
the natural genetic and phenotypic variation present in plant populations. I’m
particularly interested in genetic mechanisms responsible for key agronomic,
quality, and stress-adaptive traits that are critical to crop production in
areas prone to intense abiotic stress pressures (e.g. heat, drought. etc).
By understanding this variation and identifying
responsible ways to utilize it, we can invent solutions to global challenges
such as food security or fiber needs that our growing population is facing.
My research program is composed of three separate
but synergistic areas that combine to elucidate the genetic mechanisms
responsible for key agronomic, quality, and stress-adaptive traits.
In the first area, I try to understand the forces
that shape the differences in phenotypes we observe, disentangling the effects
of genetic variation, environment stress, and their interaction. I explore how
the genetic diversity present in plant populations contributes to phenotypic
variability, and how genes interact with the environment to give rise to
phenotypic plasticity.
The second area focuses on phenomics – the study of
phenotypes – with a special interest in traits that show temporal expression
patterns. I describe and measure variation of phenotypes (e.g. leaf size,
transpiration) in response to environmental fluctuations throughout the plant’s
life cycle.
The third area of my research revolves around the
use of statistical methods and genetic mapping populations to discover allelic
variants and causative genes that explain the phenotypic variation that I
study.
The findings from
my work offer new tools, news ideas, and new solutions to develop improved crop
cultivars that will meet the socioeconomic demands and environmental
constraints of the future.
Crops that we work on
Equipment &
Modeling
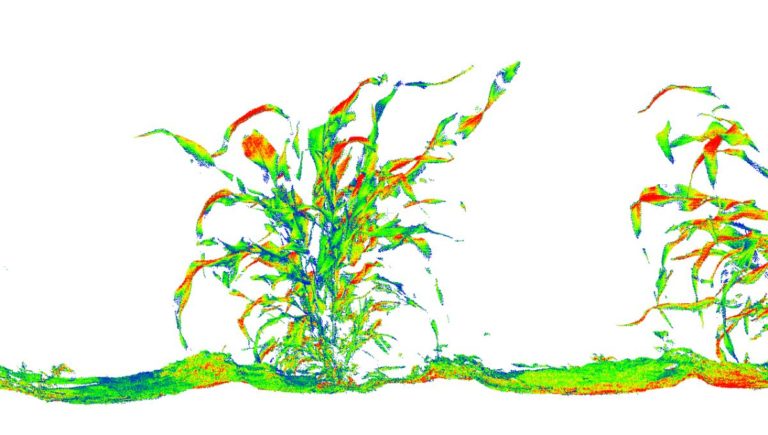
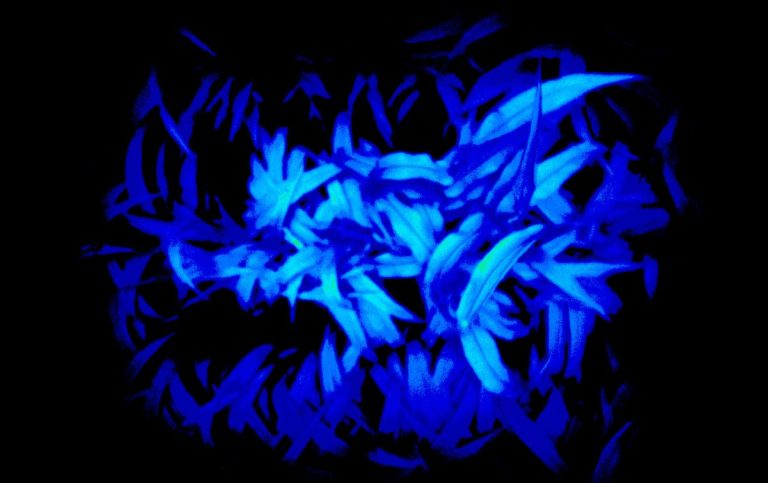
The University of Arizona (UA) Field Scanner is the worlds most advanced and complete phenotyping solution offering unparalleled insights into the plant lifecycle. The UA Field Scanner is a fully automated system that is capable of measuring crop growth and development efficiently, accurately, and quantitatively across the growing season. This longitudinal, continuous collection of high spatiotemporal resolution data enables the capture of deep phenotypes that can elucidate the mechanics and dynamics of plant development and physiology in relation to environmental conditions. To deliver these data, a suite of high-quality cameras and sensors are housed in a weather-proof cabinet that is moved over the field area by the supporting 30-ton robotic gantry. This provides accurate and consistent placement of the sensors relative to the crop canopy delivering robust and highly detailed data of unprecedented amount; two to six terabytes of data are capable of being generated per day. By coupling these cutting-edge sensors with newly developed, high-throughput data processing pipelines, traits such as ground cover, canopy height, plant geometry, growth and biomass, counting features, growth stages, vegetation indices, canopy composition, photosystem performance, and many more can be quantifiably described and recorded. Additionally, data fusion techniques are being used to integrate data collected from multiple sensors with different spatial and spectral resolutions to produced fused data sets that contain more information compared to the individual sensors. Through the integration and synthesis of these data streams, novel biological insight is being generated to help solve basic science questions as well as delivered applied solutions for the breeding and development of new crop cultivars and meeting the challenges facing a growing global population.
Phenotyping is one of the greatest limiting factors upon the rate of crop improvement. To ensure that agricultural production can increase in the face of a changing climate, it is imperative that researchers identify and implement more effective means of phenotyping plants. Emerging technologies, including remote sensing via unmanned aerial systems, or UAS, are attractive alternatives that could greatly increase the efficiency of crop improvement programs. Nonetheless, efforts must be made to determine how to implement these technologies and use the data that they generate. To that end, various high-throughput phenotyping projects are currently underway within the program. Rotary-wing UAS equipped with payloads including red, green, and blue (RGB) and thermographic, or infrared, sensors are currently in use, and these devices are being flown or will be flown over economically important crops including cotton (Gossypium hirsutum L.), sorghum (Sorghum bicolor L. Moench), and lettuce (Lactuca Sativa L.). We are using photogrammetry software and algorithms to process these data and extract phenotypes of interest. RGB data is being used to elucidate phenotypic characteristics such as canopy coverage, plant height, estimated yield, growth rate, and more. Thermographic data is being used to determine canopy temperature which is indicative of drought stress levels but also of pathogens such as fusarium wilt in lettuce. Through harnessing UAS imagery with quantitative genetics, we are working towards elucidating the genetic mechanisms that give rise to phenotypic plasticity.
Irrigation for agriculture accounts for 70% of human freshwater use. Without capacity to increase available water, this constraint poses the greatest challenge facing future food and natural resource security. Increases in plant productivity conferred by more effective use of water by plants is thereby needed. However, lack of understanding of the biophysical processes and genetic architecture of traits controlling whole-plant water movement (termed hydraulics) and their relationships and potential trade-offs to yield and quality limits development of more water efficient varieties that have the potential to reach market. Our research in the area of ecophysiological modeling addresses this need by linking genetics, phenomics and physiology via biophysical process-based models. We are leveraging expertise in field-based phenomics and quantitative genetics, genotype x environment modeling (Dr. Wang; Purdue Univ.), stress physiology (Dr.Ewers; Univ. of Wyoming), and biophysical modeling (Dr. Mackay; Univ. at Buffalo), targeting cotton (Gossypium hirsutum L.), a key species for plant-based fiber production. Our teams research aims to elucidate the genetic and physiological mechanisms that underlie their drought response by evaluating diverse germplasm under large-scale field experiments using high-throughput phenomics to develop and test biophysical plant models that simulate across different genotypes.
Meet the Team

Duke Pauli
Principal Investigator
Hello! My name is Duke Pauli and I am an associate professor in the School of Plant Sciences here at the University of Arizona. I was born and raised in Montana where I earned my PhD in Plant Sciences from Montana State University in 2014. My graduate research focused on the application and integration of genomic technologies to the breeding of improved malting barley varieties for the state’s growers. Upon completion of my PhD, I moved to Ithaca, New York where I was a Cotton Incorporated Postdoctoral Fellow in the lab of Dr. Michael Gore at Cornell University. There, my research focused on using high-throughput phenotyping (phenomics) to reveal the genetic basis of stress adaptive traits in cotton as well as how phenomics could be more broadly applied to crop improvement. Since starting my faculty position here at UA in 2018, I have been focused on developing a research program centered on understanding the genetic basis of stress adaptive traits in crop plants through various mechanisms including phenomics, field-based physiology, and quantitative approaches such as models. Broadly, I am interested in all aspects of agriculture from the historical perspective to the actual agronomic practices – I love being involved in ag!
Contact Information
dukepauli@arizona.edu
Sebastian Calleja
PhD Student
I focus on collecting both physical and physiological plant measurements at the Maricopa Agricultural Center, primarily using the LiCOR LI-6400xt portable photosynthesis system, to support the development of ecophysiological models for understanding heat and drought stress. I also aid in other tasks to support the research we do here at MAC incluidng planting, maintaining fields, and other types of data collection. I am eager to learn more about agriculture and excited to get some experience.

Jeffrey Demieville
R&D Engineer/Scientist III
I earned an M.S. in Systems Engineering from the University of Arizona and a B.S. in Biological and Agricultural Engineering from Texas A&M University. Since 2018, I’ve served as the lead engineer for the Maricopa Field Scanner Sensing Platform, originally part of the Department of Energy ARPA-E TERRA-REF project. I have done previous work in high-throughput phenotyping while I was obtaining my Bachelor’s degree at Texas A&M University as well as the International Crop Research Institute for the Semi-Arid Tropics (ICRISAT). My interests lie mainly in sensors and robotics, operations research, and reliability engineering, but my skills are often needed for many additional tasks to support the research operations at the Maricopa Agricultural Center and further afield.

Emmanuel Gonzalez
PhD Student
After graduating from Pacific Lutheran University with a bachelor’s degree in biology, I joined the University of Arizona’s Plant Science PhD program. My research interest lies in how plants sense and respond to their environment. As the world population continues to grow, arable land is diminishing. These conditions collectively threaten crop yields and food security across the globe. I hope to study emerging phenotyping technologies that could one day help accelerate plant breeding to meet growing food demands.

Clay Christenson
PhD Student
After graduating from the University of Nebraska-Lincoln with a bachelor’s degree in Plant Biology and Mathematics, I joined the University of Arizona’s Plant Science PhD program. My research interest focuses on modeling plant water hydraulics across the soil-plant-atmosphere continuum. I plan to then leverage these models in an effort to determine the genetic control of hydraulic traits which play a role in the plant’s response to drought. As the climate continues to get warmer and our access to available water diminishes, the development of crops that are less water intensive is crucial. I hope that a better understanding of genetic control of plant hydraulics can one day lead to the development of crops that are less prone to drought stress.

Bella Salter
Student Researcher
I am a sophomore at the University of Arizona majoring in computer science and mathematics, with minors in biology and statistics. I have always been interested in applying my experiences in mathematics and computer science to the field of biology, since I usually spend my free time hiking or camping. After I graduate, I aim to work towards a master’s degree related to machine learning or data science, and I hope to continue using this for biological research.

Aditya Kumar
Student Researcher
As a Computer Science major with a minor in statistics and data science, I became a member of Pauli Lab in April 2023 to expand my knowledge and skills in Data Analysis and machine learning. I am passionate about all areas of Computer Science and eager to gain valuable experience in this lab. After I graduate, I plan on pursuing at least a master's in either Computer Security or Machine Learning, but I am yet to make up my mind!

Reza (Rey) Sanayei
Student Researcher
As a senior at the University of Arizona majoring in Computer Science with a minor in Statistics and Data Science, I am deeply passionate about Machine Learning, Natural Language Processing, and Computer Vision. I am thrilled to apply these interests to the Pauli Lab undergraduate research team, focusing on Computer Vision projects. Leveraging my background in Computer Graphics and Neural Networks, I am eager to contribute to the lab's innovative work in identifying allelic variants and causative genes for phenotypic variations. Post-graduation, I aim to further my studies in these dynamic fields through graduate education.

Emily Cawley
Student Researcher
I am a photographer and a senior Computer Science and Global Media Studies major at The University of Arizona. I joined the Pauli Lab in 2023 and previously worked with the PACT Consortium. My area of interest is in AI/Machine Learning and after I graduate, I hope to work within the industry to combine my two areas of study.

Mohammad Gohardoust
Postdoctoral Researcher
I hold bachelor’s and master’s degrees in agricultural engineering with a focus on irrigation and drainage from the University of Mazandaran, Iran. My master's research involved numerical modeling of the effective unsaturated hydraulic conductivity of soils with 1-D heterogeneity. In 2015, I joined Dr. Tuller’s research lab at the Soil, Water and Environmental Science Department at the University of Arizona for my Ph.D. My dissertation focused on physicochemical characterization of soilless substrates, incorporating numerical modeling to simulate management scenarios related to container shape, irrigation, and fertigation. Post-graduation, I continued research at the University of Arizona, working on diverse projects, including collaborations with Dr. Pauli’s lab. Currently, I'm with his lab, focusing on modeling aspects of growing cotton to understand soil-plant-atmosphere interactions. I aim to apply my numerical modeling and machine learning skills to better understand the involved phenomenon.

Bryan Pastor
Research Specialist
While earning a bachelor's degree in Controlled Environment Agriculture, I started working for the University of Arizona in 2007. I worked for the Arizona Weeds Science Specialist, Dr. William McCloskey until 2022, and then with his successor, Dr. Jose Dias until September 2023 at which point I joined the Pauli lab. I have recently taken over maintaining crops and general maintenance in the gantry field. I will also run and manage the gantry as well as collecting data in our other experiments around the Maricopa Agricultural Center.

Rodrigo Silva
Masters Student
Hi there! My name is Rodrigo Silva. I was born and raised in Brazil, where I completed a program as an electromechanical technician from the Federal Center for Technological Education of Minas Gerais and earned a B.S. degree in Agronomy from the Federal University of Lavras. Before joining the Plant Science M.S. program at the University of Arizona, I worked in various industries such as automotive, fruit processing, horticulture, and fertilizer. My research interests are integrating emerging technologies into crop production to create a more sustainable and resilient agriculture. I am particularly interested in studying the stress-adaptive traits in plants, which could increase food safety around the globe. That is a project that I want to be part of.
Publications
Gonzalez, E., Zarei, A., Hendler, N., Simmons, T., Zarei, A., Demieville, J., Strand, R. Rozzi, B., Calleja, S., Ellingson, H., Cosi, M., Davey, S., Lavelle, D.O., Truco, M.J., Swetnam, T.L., Merchant, N., Michelmore, R.W., Lyons, E., and Pauli, D. 2023. PhytoOracle: Scalable, Modular Phenomic Data Processing Pipelines. Frontiers in Plant Science. 14, 1112973-1112973. doi: 10.3389/fpls.2023.1112973
Read Study
Alptekin, B., Erfatpour, M., Mangel, D., Pauli, D., Blake, T., Turner, H., Lachowiec, J., Sherman, J., and Fischer, A.M. 2022. Selection of Favorable Alleles of Genes Controlling Flowering and Senescence Improves Malt Barley Quality. Molecular Breeding. 42:59. doi: 10.1007/s11032-022-01331-7
Read Study
Thorp, K.R., Calleja, S., Pauli, D., Thompson, A.L., and Elshikha, D.E. 2022. Agronomic Outcomes of Precision Irrigation Management Technologies with Varying Complexity. Journal of American Society of Agricultural and Biological Engineers. doi: 10.13031/ja.14950
Read StudyOpportunities
We are always looking for talented, passionate people to join the lab! If you have a strong interest in quantitative genetics, plant breeding, field physiology, data science, or abiotic crop stress, please send us your information!
Postdoctoral Associates: There are currently no open positions in the lab. However, we are happy to work with you on submitting a proposal to funding agencies such as NSF Postdoctoral Research Fellowships in Biology or the USDA-NIFA Postdoctoral Program. Submission and proposal information and deadlines vary by program so it is highly recommended that interested individuals check the respective program websites on a frequent basis. If interested in applying for a fellowship or other funding mechanism, please send a CV, cover letter, and outline of your proposal to Duke Pauli at dukepauli@email.arizona.edu. Please do this well in advance of the proposal deadline so that we have adequate time to prepare a competitive proposal.
Graduate Students: Prospective graduate students should apply online at the School of Plant Sciences website (https://cals.arizona.edu/spls/graduate) for consideration of admission to the program. Although limited funding is available, prospective students are expected and encouraged to secure their own, independent funding throughout their time in graduate school. The American Society of Plant Biologists has made available a listing of federal programs and fellowships that provide support for plant science graduate students.